What's New - RiceVarMap
2026-01-29
RiceVarMap v3.0 has released: Upgrades the platform to a multi-reference genome system (Nipponbare, ZS97, MH63) and expands population genetics and deep-learning–based variant annotations. An NLP-powered query assistant and optimized architecture further enhance analytical capability and performance.
2024-09-16
Short tandem repeat (STR) data has been added to RiceVarMap2 as a type of genetic variant. It can be queried like other variant types and is available for bulk download on the download page. Special thanks to Dr. Xianrong Xie at SCAU for providing the data.
2024-02-02
On the 'Search for Variation information by Variation ID' page, users can directly access the cultivation information corresponding to each genotype through the links in the table.
2023-01-31
Fixed the error that less than 4 results could not be returned properly when designing primers. Thanks the users reported this bug.
2022-12-09
Bug fixed: Fixed the problem that some gene IDs could not be queried in the database. Thanks Dr. Haifu Tu reported this bug.
2022-07-29
RiceVarMap now offers genotype files in VCF format and Plink format for download. Please visit Data Downloads page.
2022-05-17
Rice quantitative trait nucleotides (QTNs) and inferred QTN effects were integrated into the RiceVarMap database. We thank Wei et al. for providing this very valuable information. Example
2022-03-16
RiceVarMap can be accessed using https://ricevarmap.ncpgr.cn
2021-07-16
Imputated genotype as well as phenotype data for the population can be downloaded from the download page now.
2021-06-30
 RiceVarMap V2.0 has been published in Molecular Plant.
RiceVarMap V2.0 has been published in Molecular Plant.
2020-05-10
We have developed a non-coding variation impact prediction tool in the "Regulatory Variant Prioritization" page. Users can use the variation IDs or upload a VCF file to query the predicted variation impacts in our database.
2019-05-15
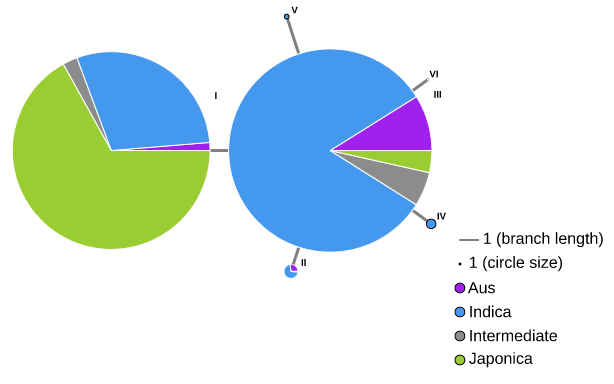
Chromatin Accessibility Map and Non-coding Varation Scores Map can be seached in "Search for Variation by Gene" page.
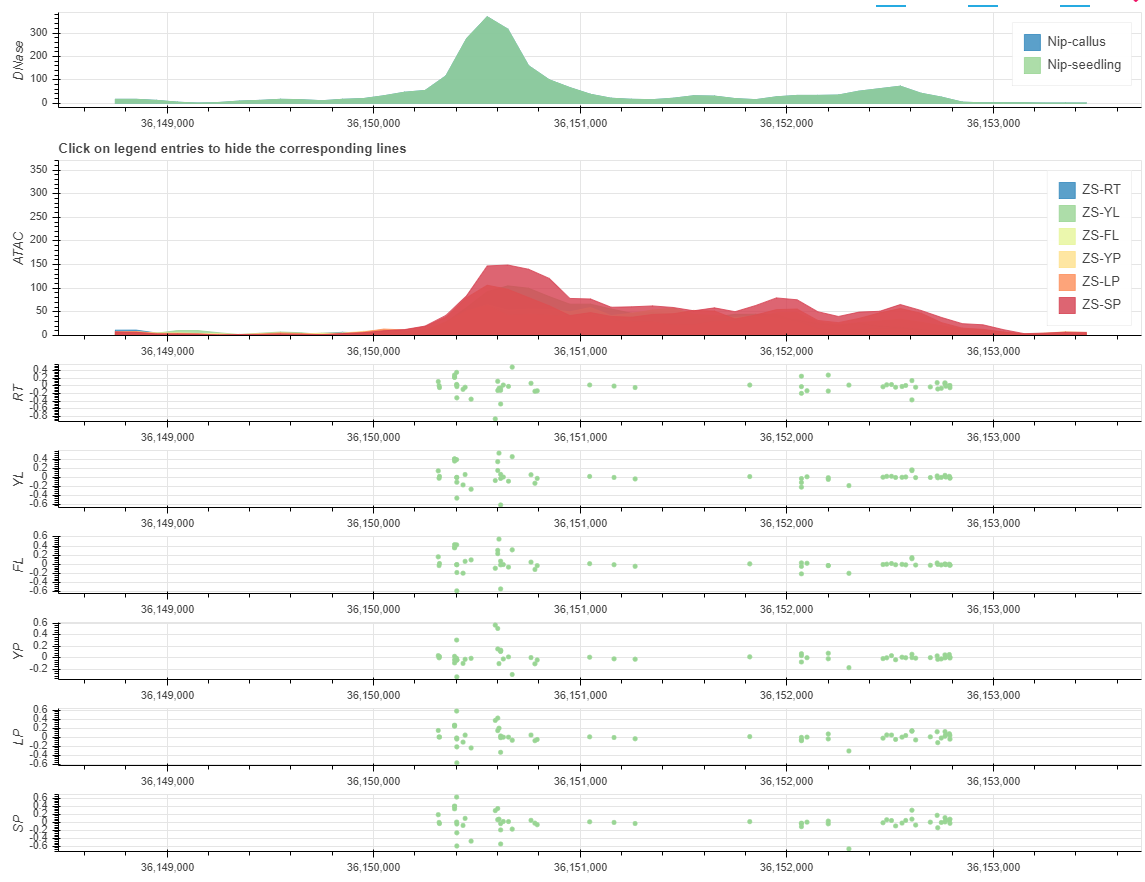
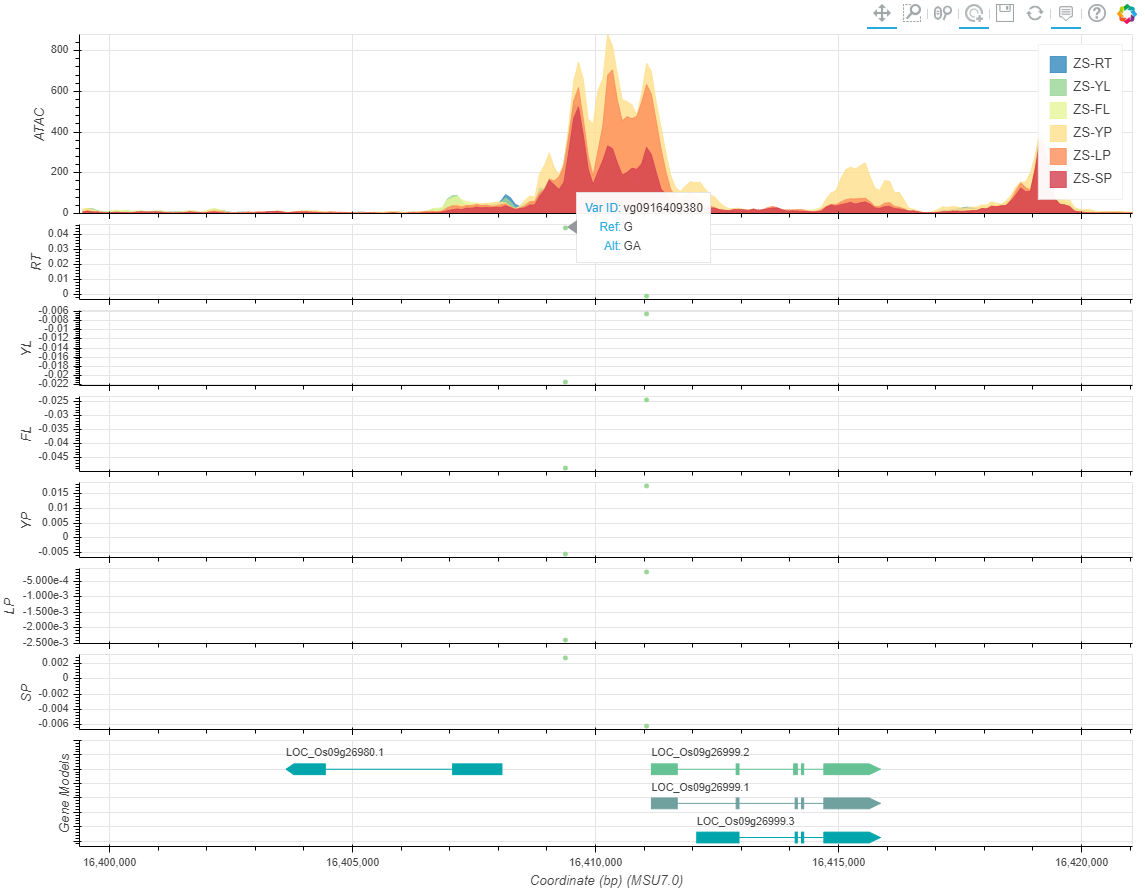
2019-04-28
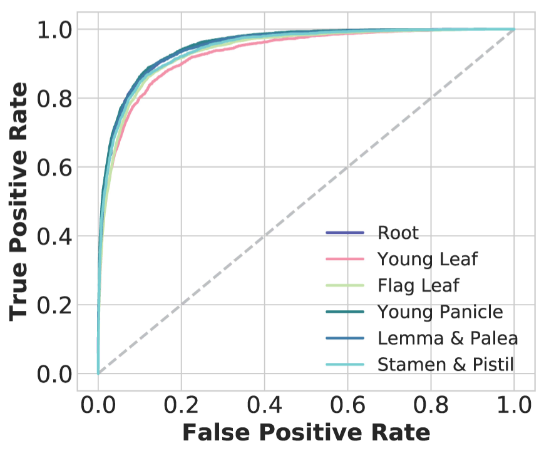
we comprehensively integrated non-coding variant prediction scores into RiceVarMap based on chromatin accessibility (CA) data and deep convolutional neural networks (CNNs).
We generated high-quality CA data from six representative rice tissues and used these data to train CNNs (AUROC: 0.93-0.95) to predict the impacts of variants on CA in non-coding regions.
2019-01-20
 We improved the speed of "Search for Genotype With Variation ID"and "Search for Cultivar Information" page.
We improved the speed of "Search for Genotype With Variation ID"and "Search for Cultivar Information" page.
2018-06-20
We defined a 'Chromatin Accessibility Score' for each variation to evaluate the impact of variations (especially for non-coding variations) based on chromatin accessibility data. This information can be found at 'Search for Variation by Gene' page and 'Search for Variation information by Variation ID' page.
2018-05-02
Gene expression atlas added into RiceVarMap, gene expression across the tissues and stages can be queried at "Search for Variation by Gene" page.
2018-04-20
We added callus, root, young leaf, flag leaf, and spikelet's chromatin accessibility data to RiceVarMap, which can be searched in "Search for Variation by Gene" page.
2017-10-08
Gene symbol and gene name like 'hwh1' can be searched in 'Search for Variation by Gene' page.
2017-09-05
RiceVarMap v2.0 has been released!
new genomic variations and GWAS results, improved annotations of missense variations, integrated chromatin accessibility data for non-coding variations.